- Home
- Resources
- Case Studies
- Decoding Neurogenetic Answers- Case Study #2
Decoding Neurogenetic Answers- Case Study #2

Patient Clinical Information: The proband is a 6-year-old female with developmental delays including speech and motor delays, subtle dysmorphic features including close set eyes, low set ears and wide space between toes, decreased IQ and staring episodes concerning for seizures.
Family History: There is no reported family history for this case.
Testing Ordered: MNG Exome™ Trio Sequencing + mtDNA
Genetic Findings: No variants that would explain the patient’s phenotype were identified through exome or mitochondrial DNA sequencing. Through copy number analysis, a heterozygous deletion of chromosome 16p11.2 was found.
Outcome: The diagnosis for this patient is 16p11.2 deletion syndrome, which is autosomal dominant. The deletion is 618 kilobases in size, encompasses 34 genes, and is consistent with other pathogenic, similarly sized deletions in this region. This variant was not detected in the patient’s parents, indicating that it is de novo.
This disorder is characterized by developmental delay, intellectual disability and speech delay. Some individuals with 16p11.2 deletion syndrome are also reported to have minor dysmorphic features, including low set ears and partial syndactyly of toes, which is consistent with the clinical history provided for this patient. Additionally, recurrent seizures have been reported, which may explain the staring episodes include in the proband’s clinical history. In this case, copy number analysis was key in determining the cause of the patient’s phenotype. Had the clinician ordered an exome without copy number analysis, they would have received a negative report.
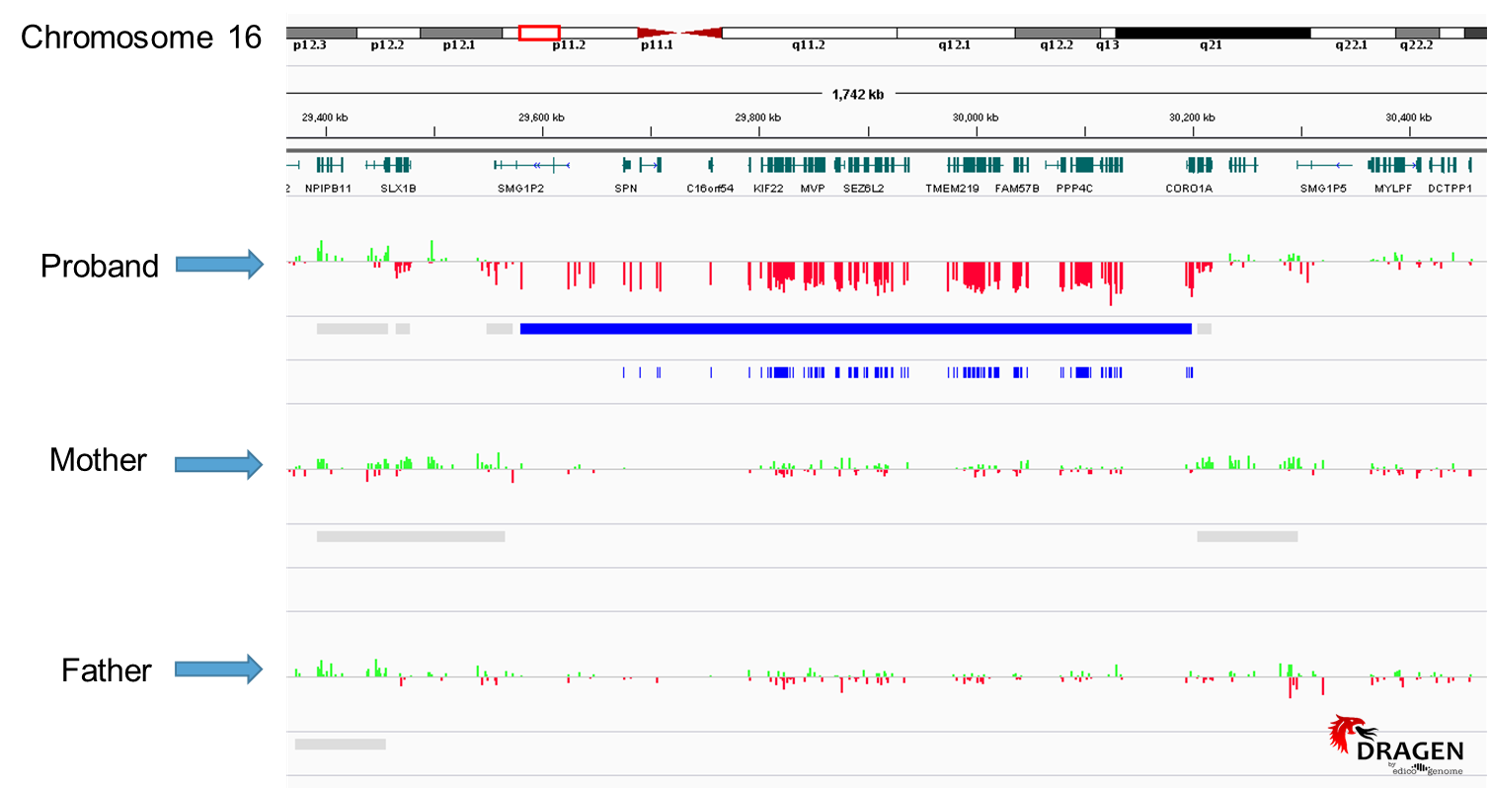
Figure1: This image is from our copy number output via the Dragen pipeline. Each red and green colored bar represents approximately 250 base pairs of sequence. The blue band shown under the proband track indicates an area consistent with heterozygous deletion, as indicated by the red bars directly above it. Directly under the solid blue bar is the zygosity track, showing apparent homozygosity for all calls in this region. This 16p11.2 deletion is clearly present in the proband, and absent in both the mother and father’s copy number data.
Reported by: Rebecca Kelly, MB(ASCP)CM

